Since I started using Raycast, I have replaced many macOS apps with it.
Here is a list of those apps and their Raycast equivalents.
Espanso
Instead of using Espanso for text expansion, I now use Raycast's built-in text snippets feature.
Hotkey
I used to use Hotkey to assign keyboard shortcuts to various actions. Now, I use Raycast's built-in hotkeys feature to achieve the same functionality.
Fly Cut
I used to use Fly Cut for clipboard management, but now I rely on Raycast's clipboard history feature to access my clipboard items quickly.
FinderGO
I used to use FinderGO to quickly navigate to specific folders in Finder. Now, I use Raycast's built-in file navigation feature to achieve the same functionality.
Rectangle
To move and resize windows, I used to use Rectangle. Now, I use Raycast's built-in window management feature to achieve the same functionality.
Textbar
Instead of using Textbar to copy TOTP for two-factor authentication, I now use Raycast's extension "One Time Password" to generate and copy TOTP codes directly.
Caffenate
To prevent my Mac from sleeping, instead of using Caffenate, I now use Raycast's built-in "Caffeinate" command to keep my Mac awake.
Calculator
Instead of using Python prompt for basic arithmetic calculations, I now use Raycast's built-in calculator feature to perform quick calculations.
This is super convenient and saves me from opening a separate calculator app.
Hammer Spoon (partially)
To assign shortcuts to browser urls, shell scripts, and other actions, I used to use Hammer Spoon.
Now, I use Raycast's built-in scripting capabilities to create custom commands and assign shortcuts to them.
Terminal (partially)
For things like killing processes, installing packages, and running shell commands, I used to rely on Terminal.
Now, I use Raycast's built-in terminal integration to run shell commands directly from the Raycast interface.
Conclusion
With Raycast, I have been able to streamline my workflow by replacing multiple macOS apps with its powerful features.

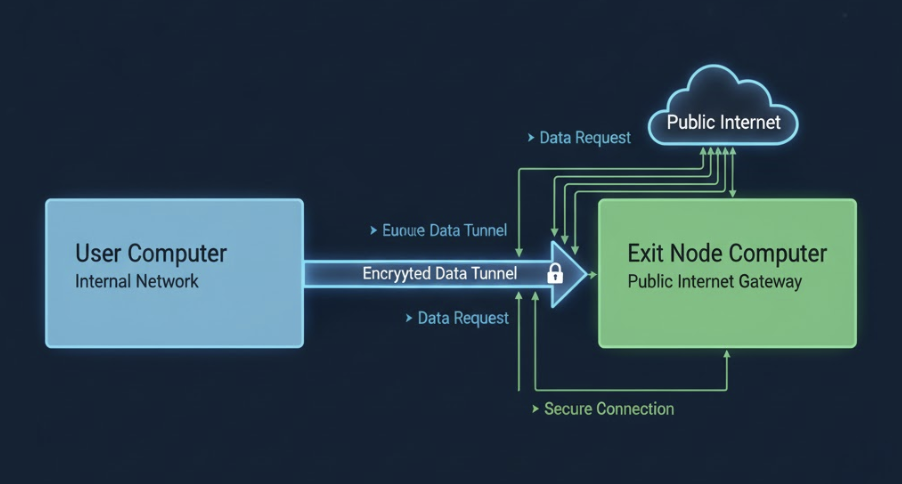
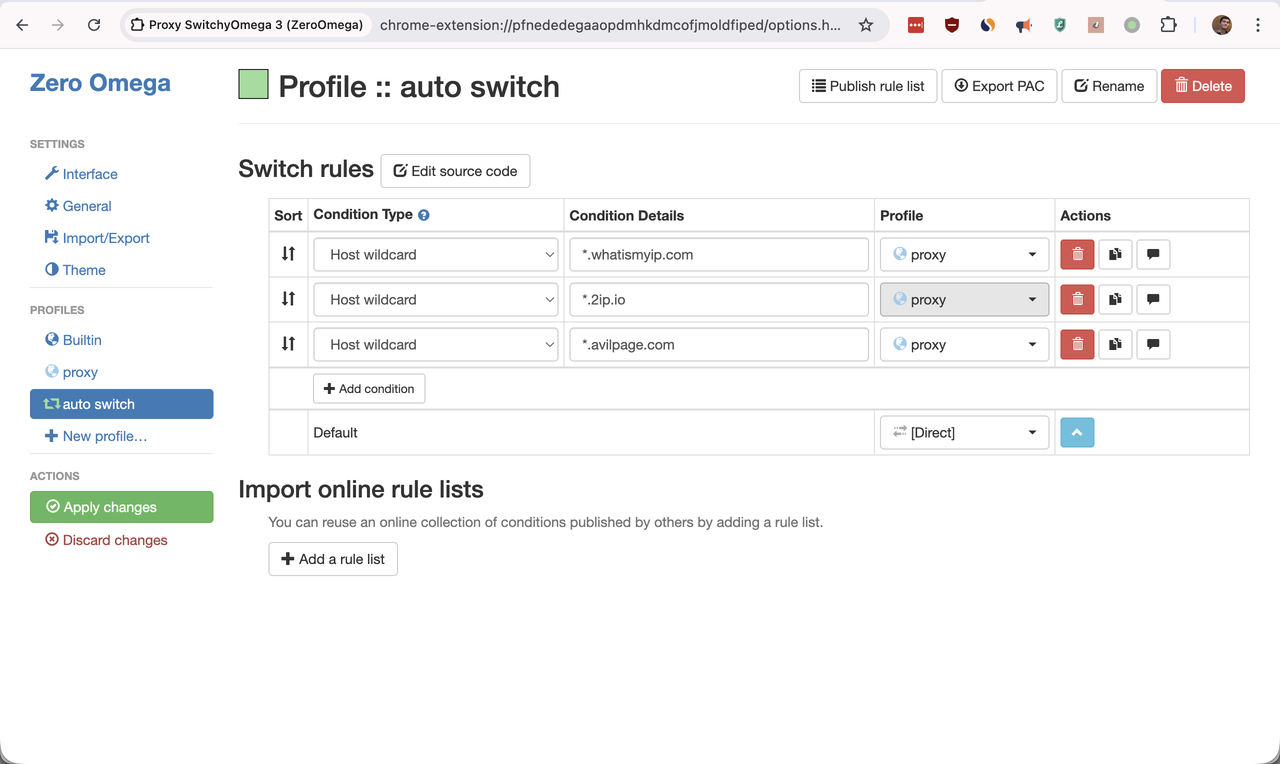
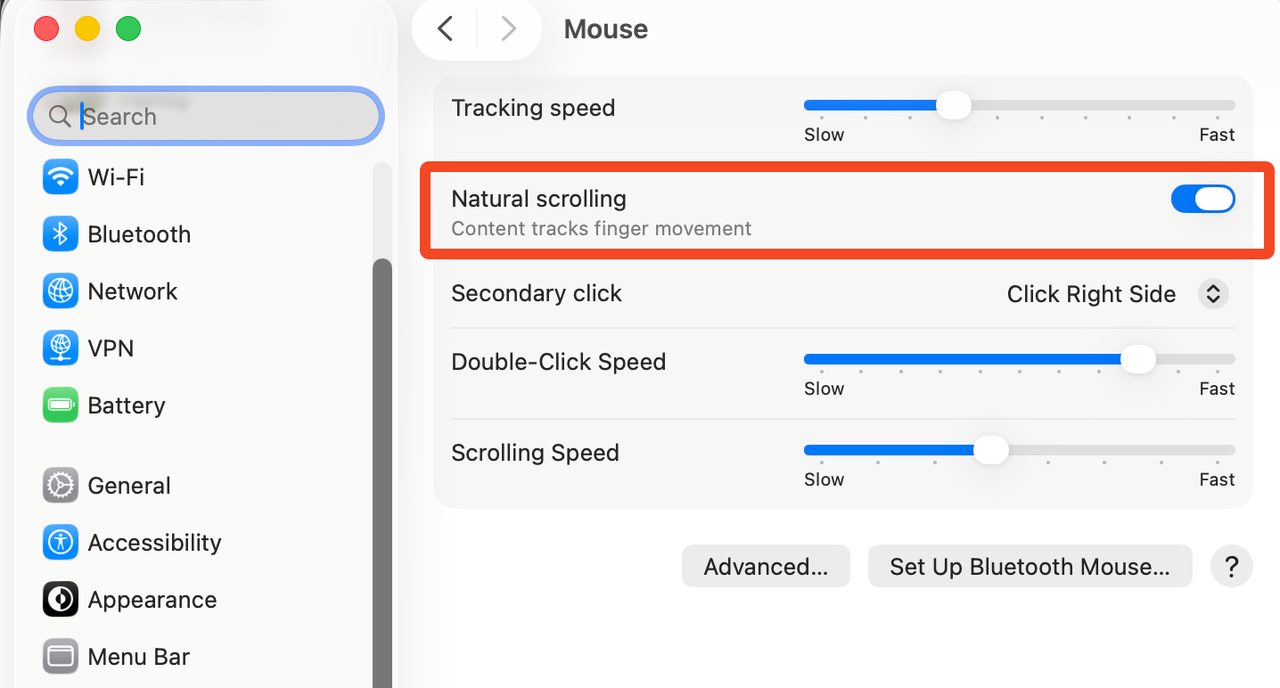
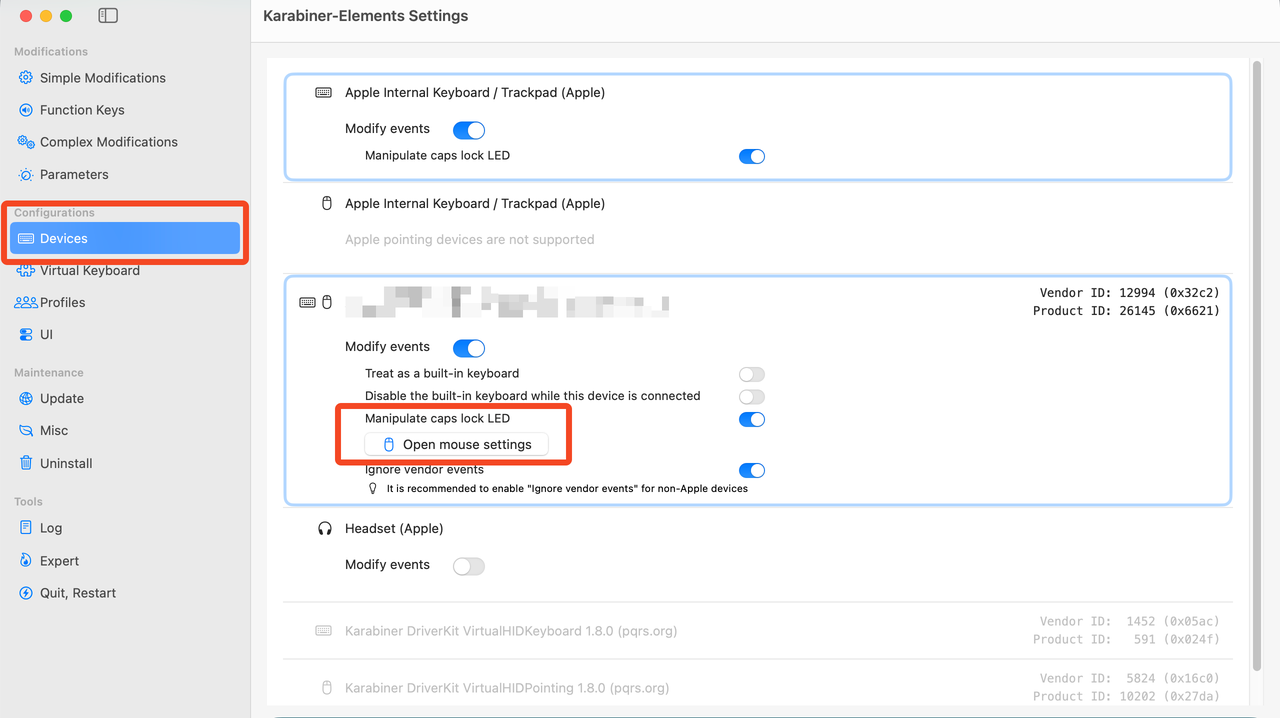
![![Raspberry Pi Imager]](../images/rpi-setup0.png)
![![Raspberry Pi Imager]](../images/rpi-setup.png)