Headlamp - k8s Lens open source alternative
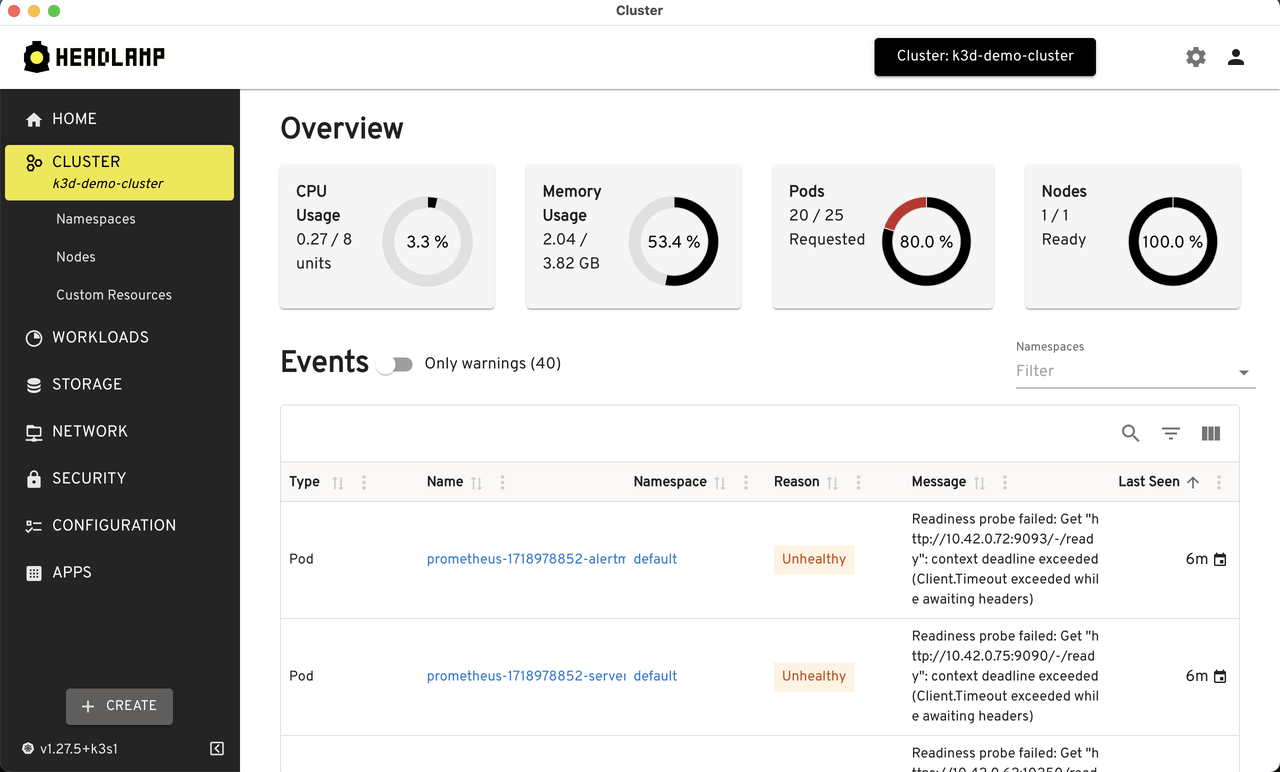
Since Lens is not open source, I tried out monokle, octant, k9s, and headlamp1. Among them, headlamp UI & features are closest to Lens.
Headlamp
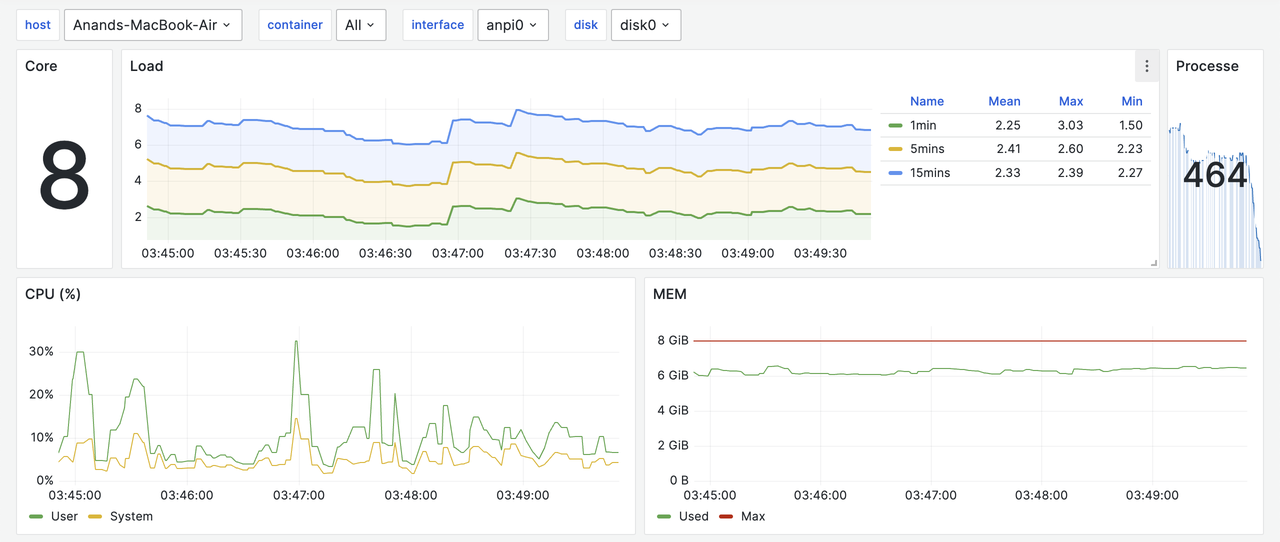
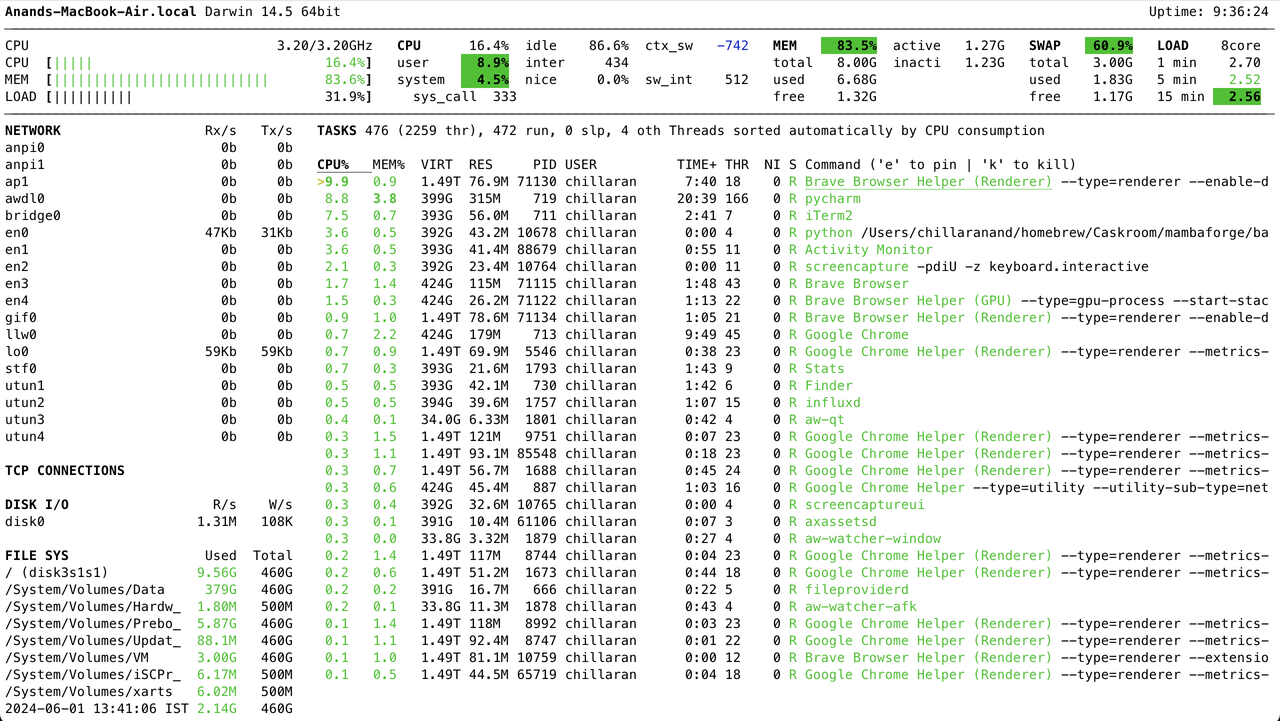
Headlamp is CNCF sandbox project that provides cross-platform desktop application to manage Kubernetes clusters. It auto-detects clusters and provides cluster wide resource usage by default.
It can also be installed inside the cluster and can be accessed using a web browser. This is useful when we want to access the cluster from a mobile device.
$ helm repo add headlamp https://headlamp-k8s.github.io/headlamp/ $ helm install headlamp headlamp/headlamp
Lets port-forward the service & copy the token to access it.
$ kubectl create token headlamp # we can do this via headlamp UI as well $ kubectl port-forward service/headlamp 8080:80
Now, we can access the headlamp UI at http://localhost:8080.
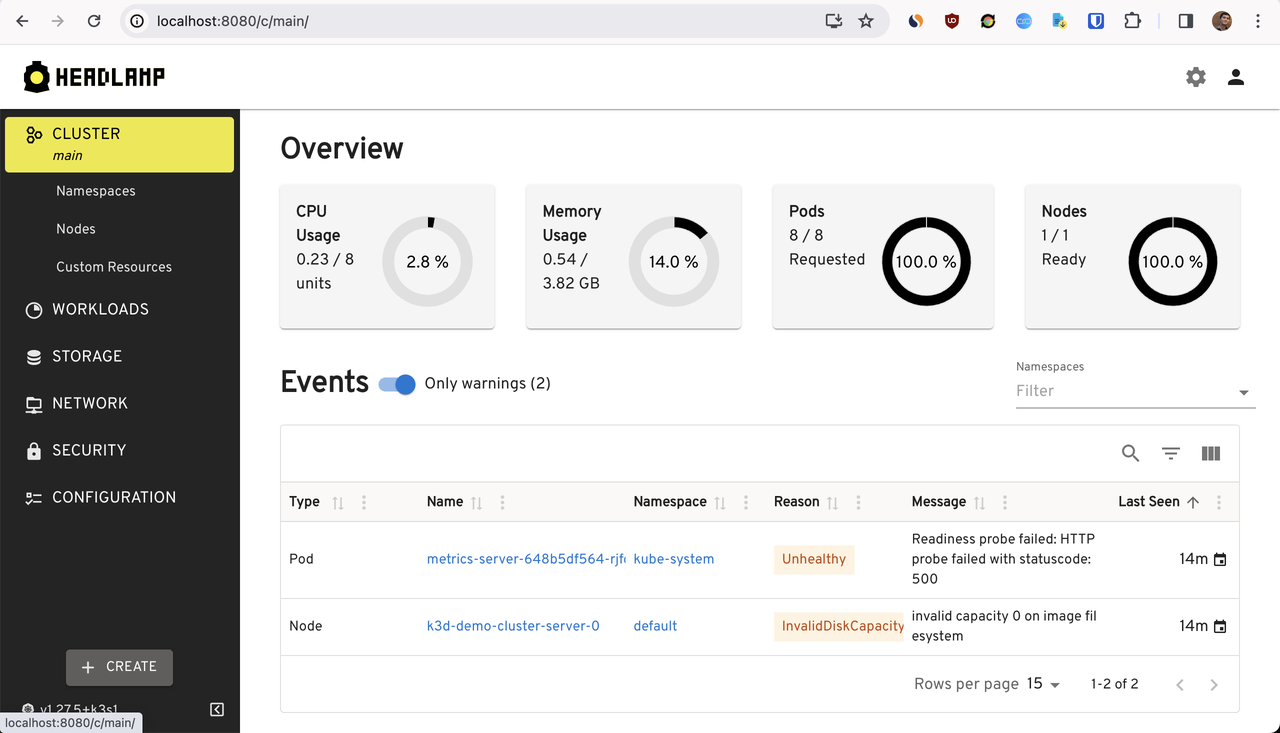
Conclusion
If you are looking for an open source alternative to Lens, headlamp is a good choice. It provides a similar UI & features as Lens and it is accessible via mobile devices as well.